CRISPR-PN2 is designed to facilitate CRISPR/Cas9 lesion experiments in parasitic nematodes
To add a nematode genome or if you have questions about CRISPR-PN2, feel free to reach out by clicking the contact button at the bottom of this page
CRISPR-PN2 returns a list of sgRNA targets as well as the starting and ending position of each sgRNA target and the strand that it is transcribed from i.e. "+" or "-"; based on this paper 20nt is the optimal length for the sgRNA: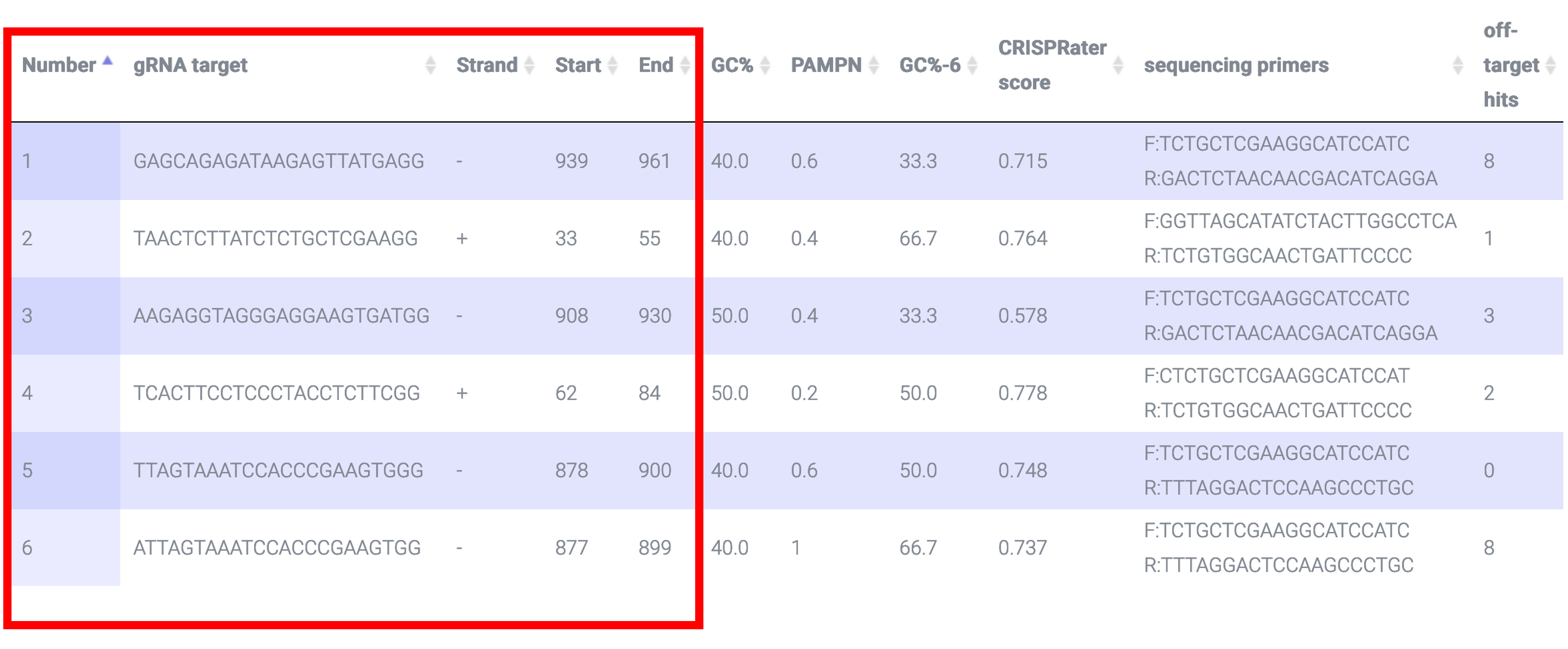
CRISPR-PN2 returns the sgRNA target GC% (i.e. the protospacer not the PAM) which should be 40-60% to maximize efficiency based on this paper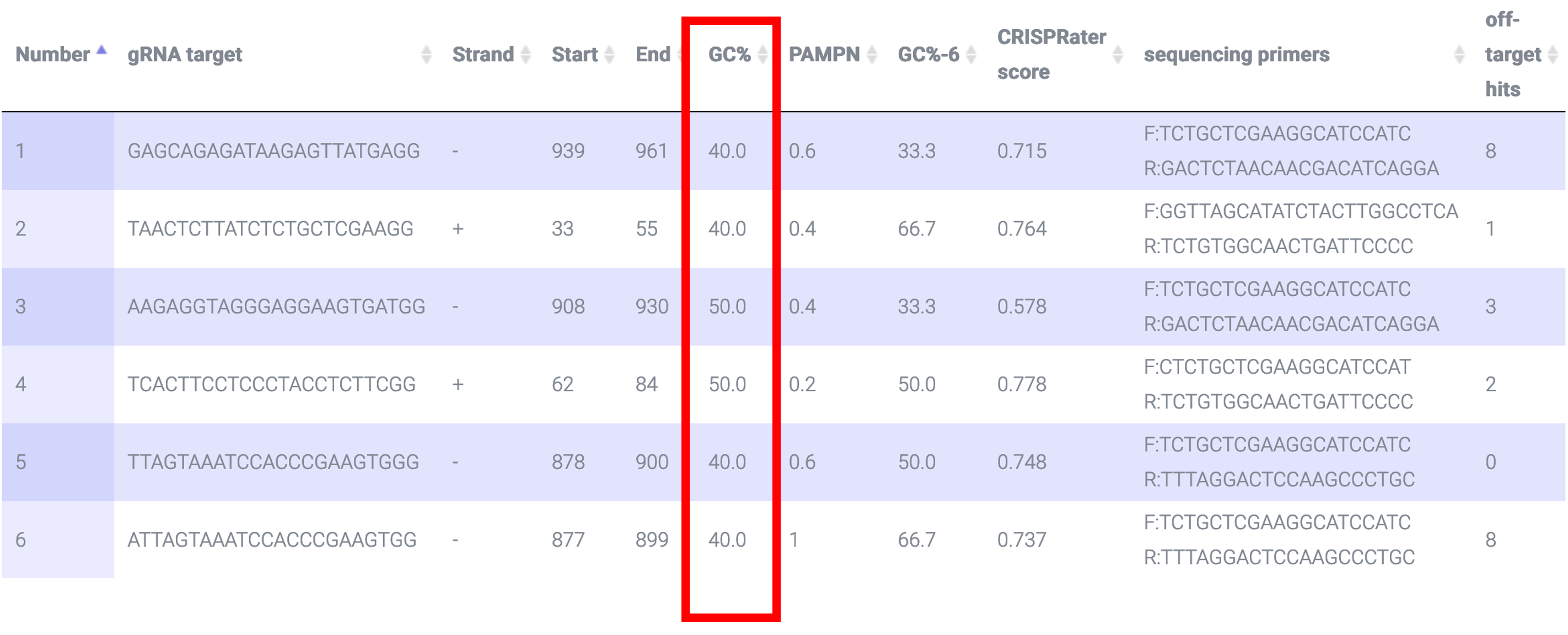
CRISPR-PN2 calculates the CRISPRater score. The CRISPRater algorithm is described in this paper. This score describes the efficacy of sgRNA such that a LOW efficacy score is <0.56, a MEDIUM efficacy score is 0.56<=score<=0.74, and a HIGH efficacy score is >0.74:
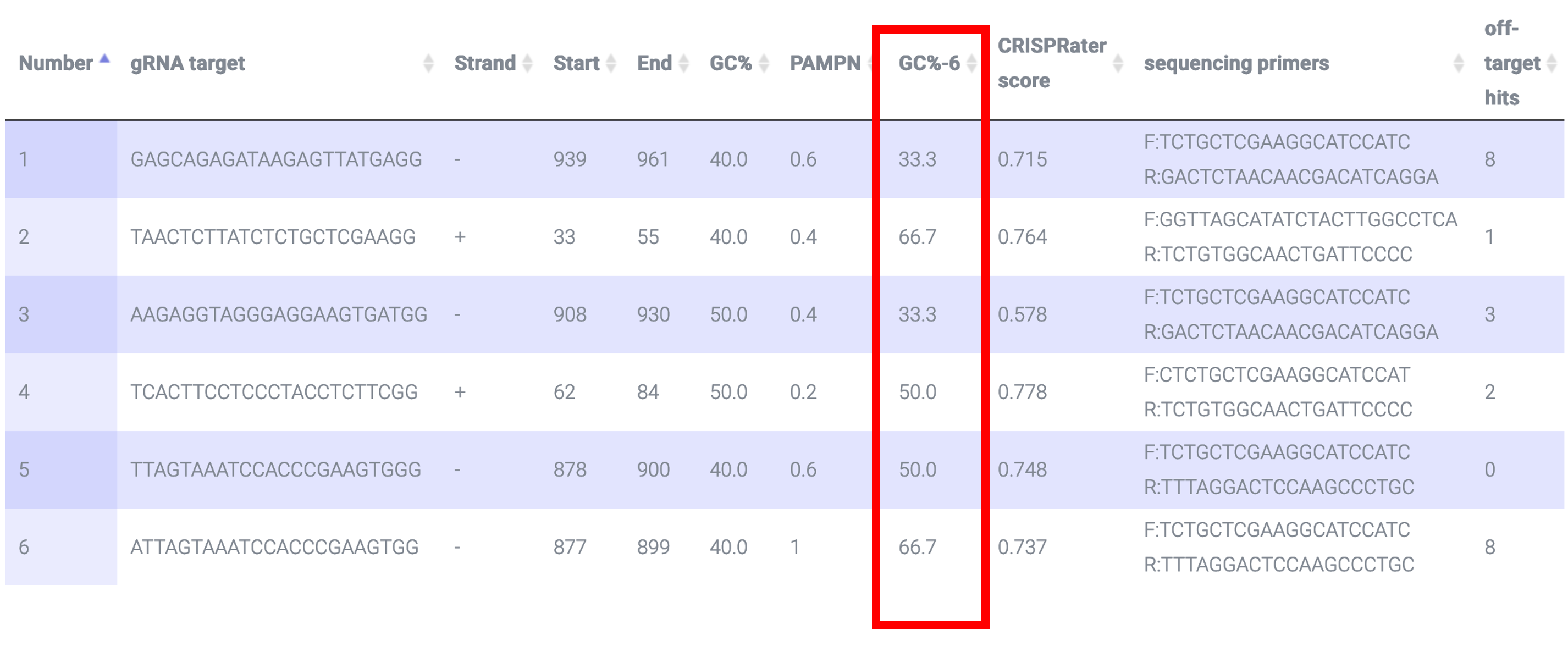
CRISPR-PN2 also returns GC% of the 6 bases immediately upstream of PAM i.e. the PAM Proximal Nucleotides (PAMPNs: green nucleotides in image below).This paper shows high GC% (>50%) in PAMPNs increase efficiency:
CRISPR-PN2 also returns a PAMPN score (max = 1, min = 0). The score is a weighted value from the four nucleotides proximal to the PAM sequence which has been shown here and here that the nucleotide position immediately adjacent to the PAM should be a "G" and not a "T" and the next three proximal nucleotides should not contain "T" in order to maximize efficiency: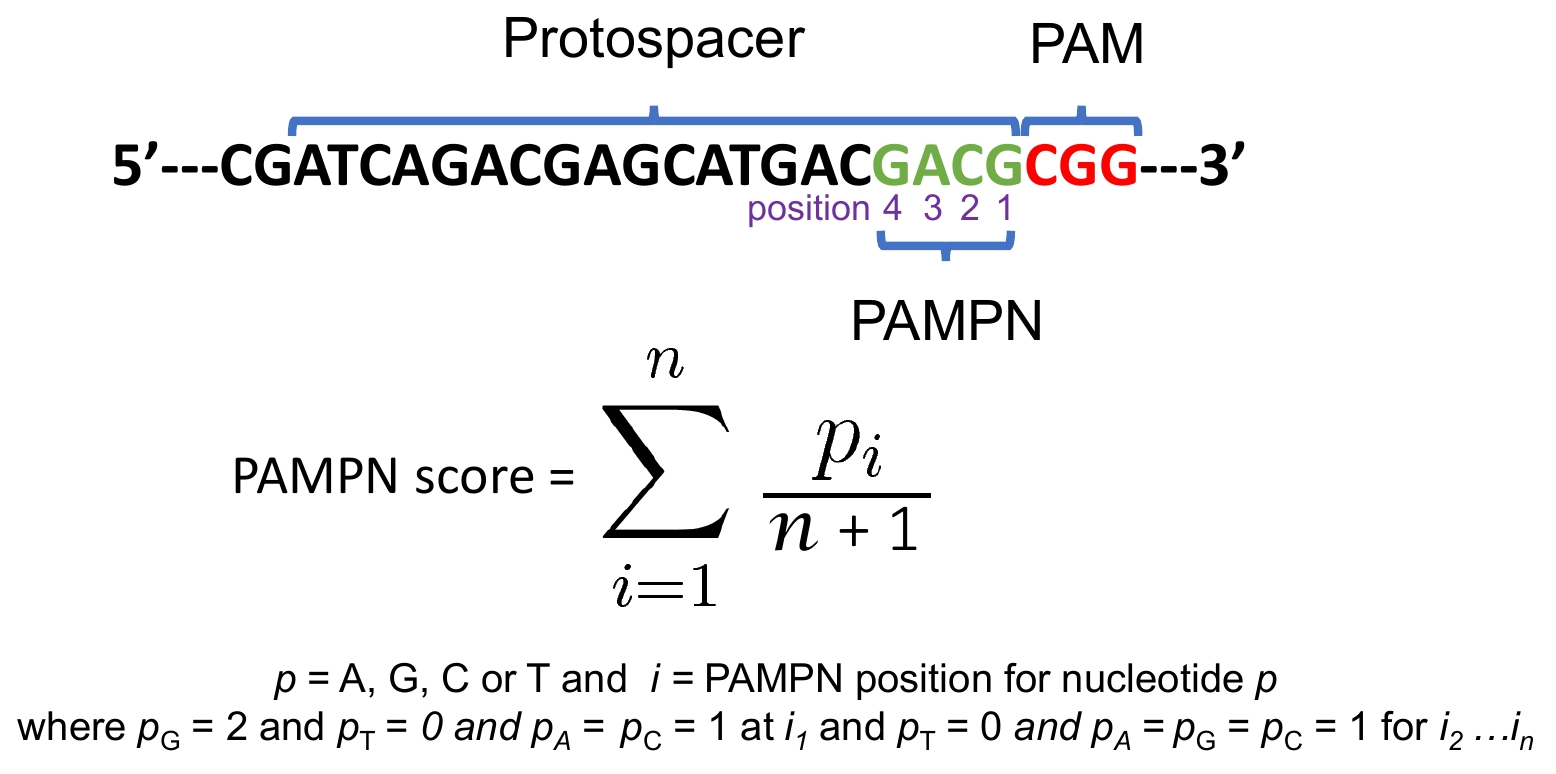

CRISPR-PN2 uses Bowtie ver. 0.12.7 to identify off-target hits, ideally with 0 mismatches in the seed region based on this paper and no more than 3 mismatches in the protospacer based on this paper: 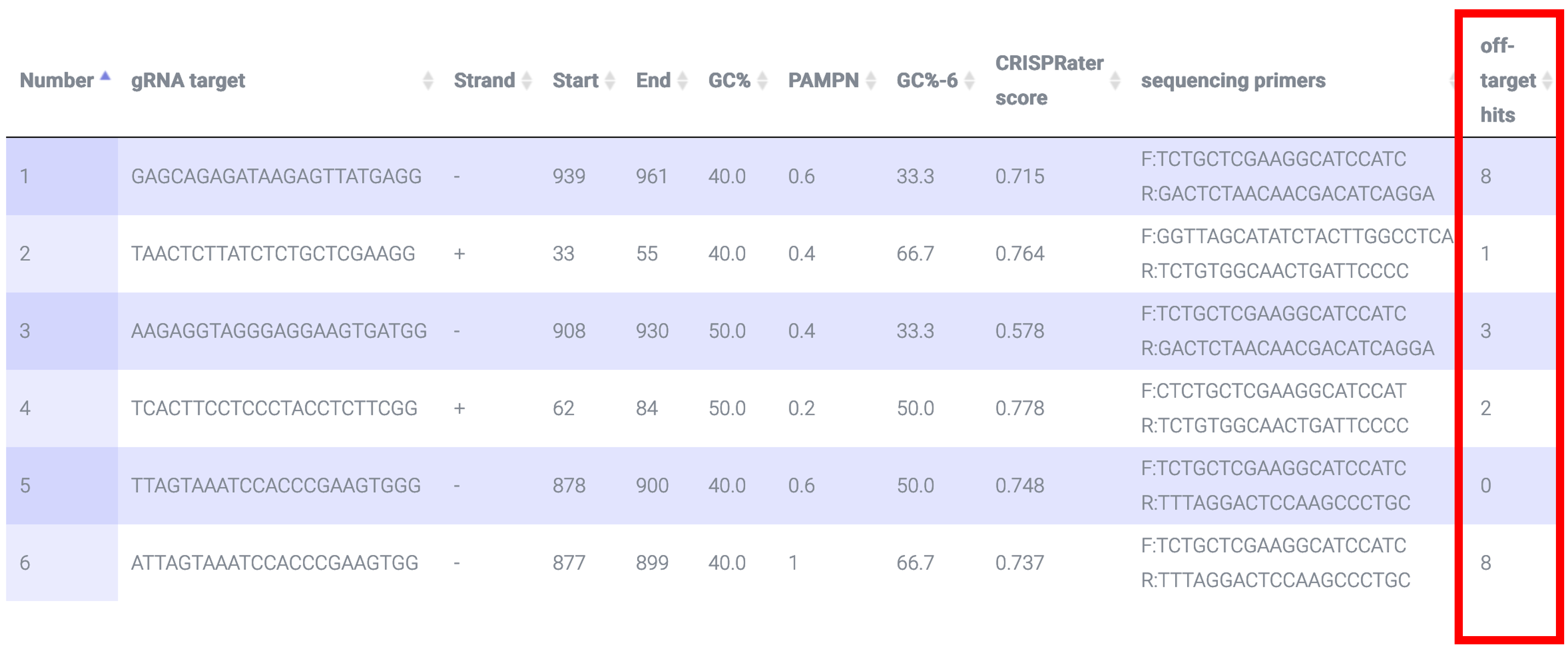
CRISPR-PN2 employs Primer3 to return sequencing primers that flank the putative lesion site: 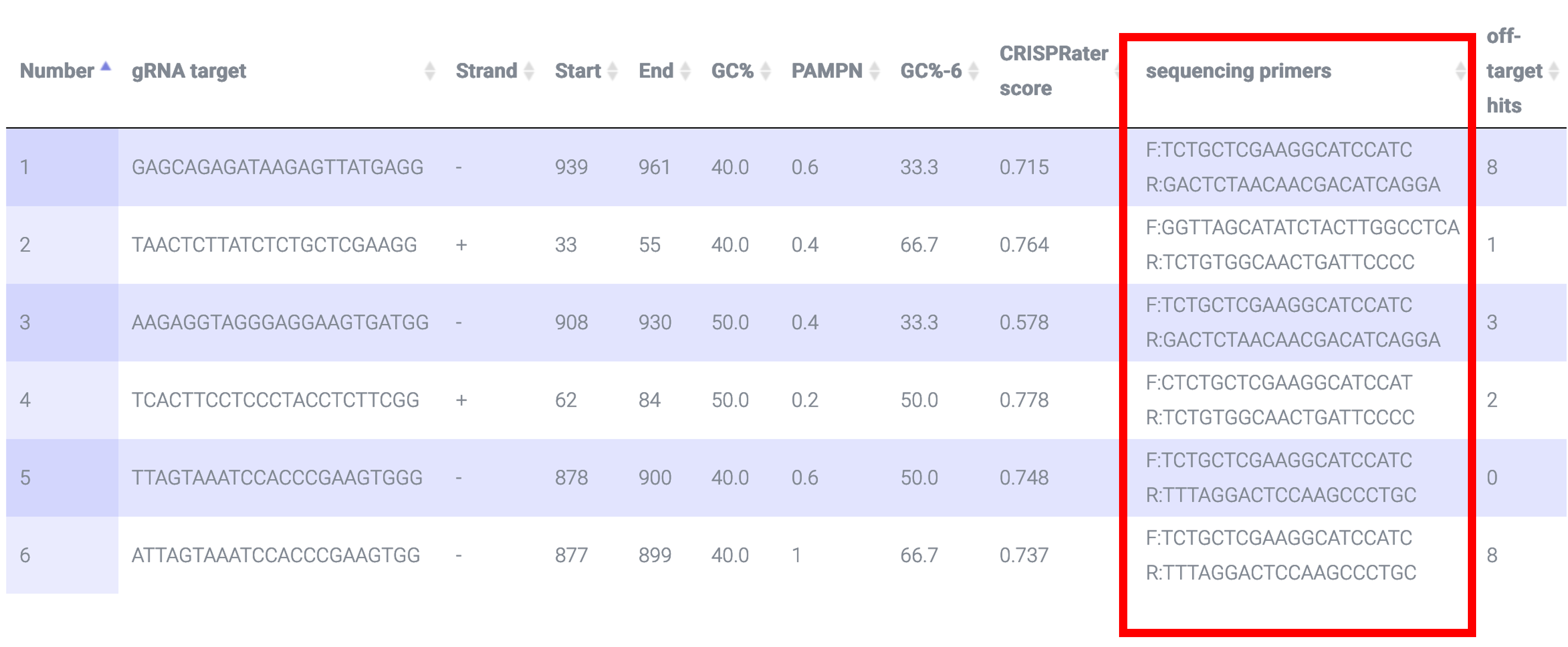
Finally, CRISPR-PN2 results include a graphical visualization of the user input sequence and sgRNA target regions rendered using Feature Viewer:
We welcome requests to add species to CRISPR-PN2. If you would like a species added, please let us know by clicking on the Contact link at the bottom of this page. Please include the species name and assembly details and we will add the new species within a few days of the request